Editorial
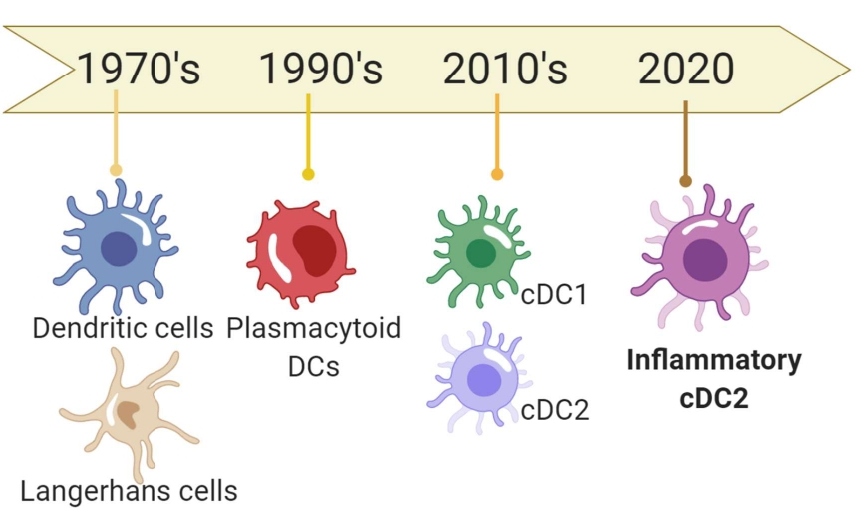
Ralph Steinman discovered and defined dendritic cells (DCs) in the 1970s [1] and at the time, no one would have expected the diversity of DC lineages to be discovered as well as their contribution to playing out the orchestra that is the adaptive immune response. In the late 20th century, researchers identified and characterized various DC subtypes, such as conventional DC (cDC), plasmacytoid DC (pDC), and Langerhans cell (LC). As DC subtypes were identified, the discoveries were followed by a systematic nomenclature. Martin Guilliams and a group of leading immunologists devised the nomenclature of DCs in 2014 which resulted in the subdivision of cDCs into type 1 cDC (cDC1) and type 2 cDC (cDC2), both of which were classified by their distinct developmental pathways as well as subsequent functions in antigen-presenting to CD8+ and CD4+ T cells, respectively [2].
Despite the extensive discoveries and characterization of these new DC lineages there is still more to be discovered. Just this year, Bart Lambrecht and Martin Guilliams, of VIB institute, elucidate new roles of cDC subtypes in Immunity. In this study, a new cDC subtype is observed, also be capable of antigen presentation and particularly under respiratory viral infection [3]. Interestingly, their study indicates that the traditional migratory and antigen presenting potential of DCs could be misclassified as roles of as monocyte-derived DCs (moDCs). Their newly discovered bona fide DC subtype, inflammatory cDC2 (inf-cDC2), is actually responsible for priming CD8+ and CD4+ T cells for Th1 responses. Specifically, those associated with viral infections and hypersensitivity induction from house dust mite, according to the report by Bosteels et al. (Figure 1).
Figure 1: A schematic timeline of DC lineage and subtype-based discoveries.
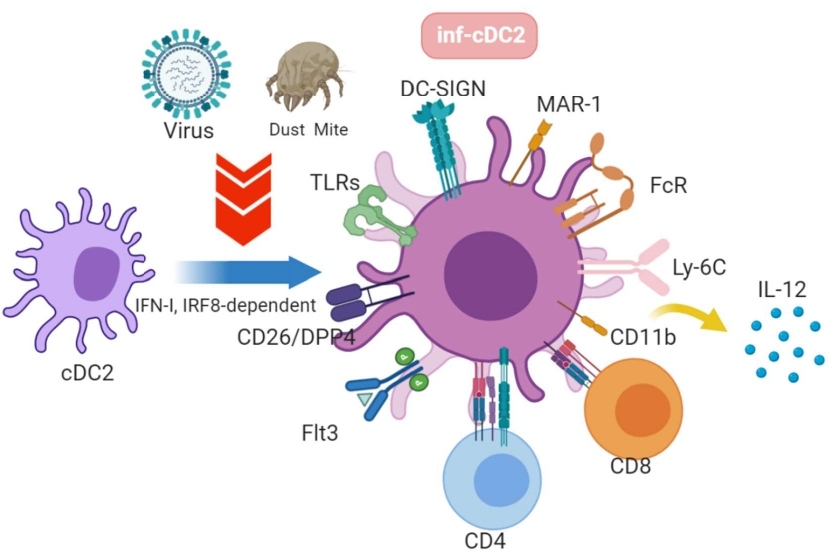
The newly discovered inf-cDC2s express cell markers that resemble parental cDC2, such as CD26, CD172a, and IRF4. They also share some features of moDCs which include cellular markers such asCD64, MAR-1, DC-SIGN, and Ly-6C. In contrast, inf-cDC2s do not express XCR-1, CD103, or CD88 cellular markers, which sufficiently discriminates them from another DC subtype like moDCs. Also, under viral infection conditions, CCR7 expression is upregulated on inf-cDC2s and required for the migratory capability of inf-cDC2s. Once matured inf-cDC2s driven by Type 1 Interferon and IRF8 signaling arrive in the infected tissues like lungs and dermis, they uptake antigens mediated by Fc receptors and present to CD4 and CD8 via MHC-II and MHC-I, respectively, to prime adaptive immunity. Inf-cDC2s can also subsequently secrete cytokines to initiate stronger immune responses (Figure 2).
Figure 2: cDC2s in steady state are developed into inflammatory cDC2 upon TLR-activation by viral or dust stimulation depending on type 1 interferon and IRF8-dependent signaling. Activated inf-cDC2s migrate to the infected tissues, uptake antigens via Fc receptor, present antigens to CD4 and CD8 T cells, and produce IL-12 cytokine to initiate the cascade immune responses.
Immunologists have been dedicated to investigating the detailed functional facets of moDCs, such as T cell priming, antigen uptake, and IL-12 production; subsequently the learnings have been transferred and applied throughout the field. Historically, moDCs were thought to be originated from common monocyte progenitors [4] and respond to the stimulation of granulocyte-macrophage colony-stimulating factor (GM-CSF) for maturation and subsequently differentiate into CD11c+ MHC-II+ moDCs in mice. It was then thought that the cells present antigens and induce T cell activation under inflammatory conditions. However, moDCs appear to be comprised of a heterogeneous population of cells expressing both of CD11c and MHC-II markers [5]. This observation may suggest that there could be more sublineages of DC’s of cells that need to be further identified. Also, the process of differentiation and the mobilized capability of moDCs seem to be enhanced during infections, notably influenza virus infection [6]. However, the findings from Bosteels et al. demonstrate another argument that inf-cDC2 cells have been misidentified as moDCs due to overlap in cellular markers and moDCs actually exhibit less migratory behavior, poor antigen presentation, and are not stimulated by GM-CSF but rather Flt3L.
Contemporary single-cell multiomics technologies, such as single-cell RNA sequencing (RNA-seq), enable researchers to insightfully examine the cellular profile of individual cells in response to the stimulation from the environment [7,8]. Single-cell transcriptomics approaches are especially handy when characterizing a group of cells for heterogeneity. More recently, single-cell RNA-seq was extensively exploited for classifying immune cell subtypes in lieu of marker-based assays. Single-cell RNA-seq could more comprehensively and unbiasedly assess the profiling of DC subtypes and their functions as described in a previous report in 2017, where they analyzed blood DCs and monocytes and pinpointed six distinct DC and four monocyte subtypes in steady state via deep sequencing [8]. As if providing the missing complementary puzzle piece, Bosteels et al. determined that inf-cDC2 cells in the viral infection state, maintain some functions similar to monocytes, macrophages, and conventional DCs during inflammatory conditions. As a result, the unique cellular markers of inf-cDC2 cells stand out when providing a higher resolution classification of DC subtype through single-cell technologies, discriminating inf-cDC2s from the conventional moDCs under the setting of viral infection, such as pneumonia virus mice (PVM) and influenza virus, as well as inflammation due to house dust mite.
While the study accomplished by Bosteels et al. has brought about a much clearer picture of DC subtypes in response to viral infections and hypersensitivity, there is still interest in whether other respiratory viruses like coronaviruses and those not causing respiratory illness could trigger this formation of inf-cDC2s as well as in how to manipulate the functions of inf-cDC2s as a target to induce stronger immunity after vaccination or to alleviate the aberrant immune responses in autoimmune diseases. Nevertheless, it appears to be necessary to better refine the functions of DCs with capability of APC and to more precisely (or at least include a CD26-based staining) depict what exact subtype of DCs playing a role in the defensive or pathogenic immune responses.
References
2. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nature Reviews Immunology. 2014 Aug;14(8):571-8.
3. Bosteels C, Neyt K, Vanheerswynghels M, van Helden MJ, Sichien D, Debeuf N, et al. Inflammatory type 2 cDCs acquire features of cDC1s and macrophages to orchestrate immunity to respiratory virus infection. Immunity. 2020 May 8.
4. Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nature Immunology. 2013 Aug;14(8):821-30.
5. Helft J, Böttcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+ MHCII+ macrophages and dendritic cells. Immunity. 2015 Jun 16;42(6):1197-211.
6. Nakano H, Lin KL, Yanagita M, Charbonneau C, Cook DN, Kakiuchi T, et al. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nature Immunology. 2009 Apr;10(4):394-402.
7. Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Frontiers in Genetics. 2019 Apr 5;10:317.
8. Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017 Apr 21;356(6335).