Abstract
DNA polymerase epsilon subunit 2 (POLE2) is a vital component of the DNA polymerase epsilon complex, essential for leading-strand DNA synthesis and maintaining genomic stability. Recent cancer studies have highlighted POLE2's oncogenic importance, as it is overexpressed in breast, lung, liver, and kidney cancers. Elevated POLE2 levels often correlate with poor overall and relapse-free survival, though some cancer types show contradictory associations, suggesting context-dependent roles. POLE2 is closely associated with the tumour microenvironment (TME), where higher levels link to lower immune and stromal scores, indicating an immunosuppressive TME. Additionally, POLE2 expression positively correlates with CD4+ memory and follicular helper T cells, while showing negative associations with regulatory T cells (Tregs), monocytes, mast cells, and M1 macrophages—highlighting its potential role in immune modulation. POLE2 also shows strong associations with immune checkpoint genes such as PD-L1 and CTLA-4, indicating a role in immune evasion mechanisms. Functional analyses further reveal its involvement in critical cellular pathways, including DNA replication, chromosomal segregation, cell cycle regulation, and DNA repair processes like mismatch repair and p53 signaling. Moreover, POLE2 is positively correlated with tumour mutational burden (TMB) and microsatellite instability (MSI), both of which are emerging biomarkers for immunotherapy responsiveness. Collectively, these insights position POLE2 as a promising prognostic and immunological biomarker with potential therapeutic relevance across multiple cancer types, warranting further in-depth investigation.
Keywords
Pan-cancer, Oncogenesis, POLE2, DNA polymerase, Tumour, Biomarker, Tumour microenvironment
Introduction
Cancer is still one of the most deadly and complicated illnesses, and research into the molecular factors that influence its progression and resistance to treatment is becoming more and more important. A crucial part of the DNA polymerase epsilon complex, DNA polymerase epsilon subunit 2 (POLE2) is essential for keeping the genome stable and helping to create the leading strand of DNA during replication [1]. POLE2 operates as an accessory component in biology, supporting the complex's repair and proofreading capabilities while stabilizing the catalytic core [1,2]. The importance of POLE2 and its associated polymerase complex in cancer lies in their crucial function of preserving genomic stability [1,3]. In particular, the lagging strand during DNA replication is synthesized in large part by DNA polymerase epsilon. If POLE2 is not working properly or is not expressed enough, it can cause problems with DNA stability, make DNA replication less reliable, and speed up tumour growth. POLE2 is a potential biomarker for prognosis and a target for therapy, as new research indicates that abnormal expression of the protein may have carcinogenic implications in various cancers.
Despite POLE2's historical recognition for its function in DNA replication and repair, new pan-cancer studies indicate a more extensive carcinogenic effect. Poor prognosis and advanced disease stages are correlated with aberrant overexpression of POLE2 across a variety of tumour types, suggesting that POLE2 plays a role in tumour growth. In addition to its normal replication function, POLE2 seems to have an impact on the tumour immune microenvironment. It may be involved in immune evasion because it coincides with higher expression of immune checkpoint genes and has a negative relationship with immunological and stromal scores. These findings suggest that more studies should be done on POLE2 as a possible target for immunotherapy, since they hint at its important but not well-understood role in managing immune responses and increasing tumour aggressiveness.
POLE2 in Tumorigenesis: Insights from the Pan-Cancer POLE2 Study and Beyond
Methodology and key datasets
The recent study used The Cancer Genome Atlas (TCGA) as its main data source, which offers comprehensive genomic, transcriptomic, epigenomic, and proteomic profiles for more than 20,000 tumour and matched normal samples covering 33 cancer types [4]. TCGA enabled a thorough pan-cancer analysis of POLE2 expression, survival results, immune infiltration, and molecular features. To provide precise comparisons between normal and cancerous states, POLE2 expression was evaluated in a variety of healthy tissues using the GTEx (Genotype-Tissue Expression) database [5]. To study DNA methylation, immune cell presence, and clinical relationships, other tools like UALCAN and TIMER were used, building a complete bioinformatics system to evaluate how POLE2 works in cancer biology [6,7].
Expression patterns and diagnostic relevance of POLE2
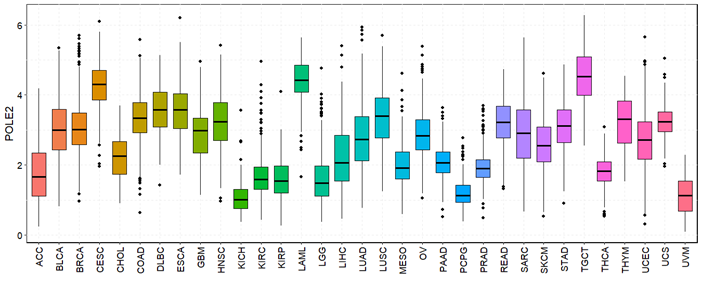
POLE2 was found to be significantly overexpressed in a majority of tumour types across TCGA datasets. Expression-level distributions are visualized using box plots utilized TCGA plot R package (v4.4.1). Elevated expression levels were particularly prominent in most of the cancers such as breast (BRCA), liver (LIHC), lung (LUAD and LUSC), and kidney (KIRP and KIRC) (Figure 1). This widespread upregulation suggests that POLE2 may act as a common oncogenic factor across diverse cancers. By comparing tumour samples with matched normal tissues, it became evident that POLE2 expression is consistently higher in malignant tissues, supporting its potential as a universal biomarker for tumour detection and progression.
ROC analysis showed high AUC values (>0.90) for POLE2 as a diagnostic marker in cancers like BRCA and LUSC. These findings position POLE2 as a robust biomarker for cancer detection.
Figure 1. The expression level of POLE2 in 33 different types of cancers in TCGA datasets. ns: not significant; *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.0001.
Survival analyses and prognostic relevance
The prognostic significance of POLE2 expression across various cancer types was thoroughly assessed using Kaplan–Meier survival analysis and Cox proportional hazards regression models. The results revealed that high POLE2 expression is significantly associated with poor overall survival (OS) and relapse-free survival (RFS) in several malignancies, suggesting its potential role as a negative prognostic marker.
For overall survival, cancers such as bladder urothelial carcinoma (BLCA), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), and sarcoma (SARC) demonstrated a strong association between elevated POLE2 expression and shorter patient survival. These observations imply that POLE2 may drive tumour aggressiveness and contribute to poor clinical outcomes in these malignancies.
Interestingly, in contrast to the general trend, a subset of cancers—such as cervical squamous cell carcinoma (CESC), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), ovarian cancer (OV), and thymoma (THYM)—exhibited a paradoxical relationship where higher POLE2 expression correlated with better overall survival. This variation suggests that POLE2’s prognostic impact is likely influenced by tumour-specific biology, including unique microenvironmental interactions, mutation profiles, or differential gene expression patterns.
Regarding relapse-free survival (RFS), a similar pattern emerged. High POLE2 expression was significantly associated with shorter RFS in cancers like KIRP, LUAD, PAAD, SARC, thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC), reinforcing its role in promoting recurrence and resistance. Conversely, in KIRC, higher POLE2 levels were linked to longer RFS, again highlighting the complexity of POLE2’s function depending on cancer type.
Collectively, these survival analyses underscore POLE2's prognostic importance. Its consistent association with unfavorable outcomes in many cancers supports its use as a potential biomarker for risk stratification.
Highlights on POLE2 and tumour microenvironment correlations
One of the most compelling aspects of our pan-cancer analysis of POLE2 is its significant relationship with the tumour microenvironment (TME), a complex and dynamic system composed of immune cells, stromal elements, signaling molecules, and extracellular matrix components. The TME not only supports tumour progression but also plays a critical role in immune escape and therapy resistance [8].
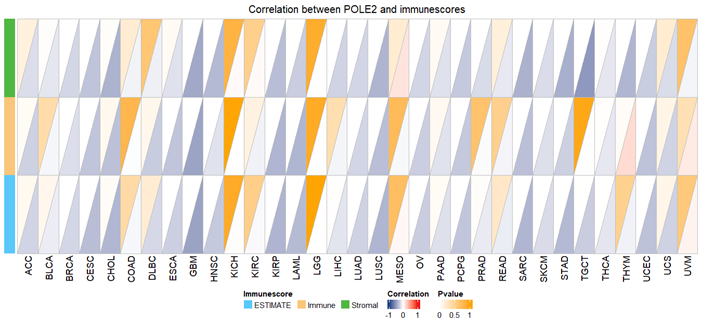
Figure 2 illustrates the correlation between POLE2 expression and three immune-related scores, such as ImmuneScore, StromalScore, and ESTIMATEScore—across multiple cancer types, calculated using the ESTIMATE algorithm [9]. Pearson correlation analysis was applied, and the results are displayed in triangular plots, where color intensity reflects the strength and direction of correlation and the shading indicates the significance level (p-value). A consistent observation was that high POLE2 expression correlates negatively with ImmuneScore, StromalScore, and ESTIMATEScore in several tumour types, including glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), stomach adenocarcinoma (STAD), and lung adenocarcinoma (LUAD). This inverse relationship suggests that tumors with elevated POLE2 expression tend to have lower infiltration of immune and stromal cells, thereby lowering the overall ESTIMATE score. Since ImmuneScore reflects the extent of immune cell infiltration, and StromalScore reflects stromal content, a reduction in both implies that high POLE2 tumors are generally associated with diminished immune surveillance and stromal contribution within the tumour microenvironment (TME) [10]. From a biological perspective, this finding highlights a potential role of POLE2 in shaping an immunosuppressive microenvironment. A reduced immune and stromal presence may provide tumour cells with an advantage by allowing immune evasion, promoting tumour progression, and reducing the effectiveness of immunotherapy [11]. Importantly, the consistent negative association observed across distinct tumour types indicates that this effect of POLE2 is not cancer-specific but may represent a general mechanism of tumor-immune interaction.
Figure 2. Correlation between POLE2 and immune score displayed by triangle. Pearson correlation was applied.
Additionally, POLE2 affected immune cells in a complex way, showing positive relationships with CD4 memory T cells and follicular helper T cells, but negative relationships with Tregs, monocytes, resting mast cells, and M1 macrophages. Tregs are essential for lowering anti-tumour responses and preserving immunological tolerance [12]. Their presence in the tumour microenvironment is frequently associated with immune suppression and tumour development. One reason why POLE2 and Tregs often don't go together in many tumors might be that POLE2 works differently than usual immune-suppressing processes. Another possibility is that tumors that are immune-excluded have high levels of POLE2 expression [13]. Resting mast cells can inhibit T cell activation, whereas monocytes can develop into tumour-associated macrophages [14]. POLE2's inverse relationship with these cells could imply a TME with reduced innate immune recruitment. On the other hand, CD4+ memory T cells are essential for maintaining long-lasting immune responses and promoting the activity of CD8+ T cells. Their presence is often associated with improved immune surveillance and long-term tumour control [15]. Additionally, Follicular Helper T Cells (Tfh) help B cells grow and make antibodies in special areas called tertiary lymphoid structures (TLSs). The presence of Tfh cells has been linked to increased anti-tumour immunity and responsiveness to checkpoint inhibitors [16]. It is important to note that the expression of POLE2 in tumors such as bladder cancer (BLCA), lung adenocarcinoma (LUAD), and uterine corpus endometrial carcinoma (UCEC) was found to be positively related to several immune checkpoint genes (like PD-L1 and CTLA4), suggesting it may be involved in helping tumors avoid the immune system. These connections imply that POLE2 could influence the effectiveness of immunotherapies and might serve as a marker to categorize the immune environment in various cancers.
Functional and molecular insights into POLE2
Gene co-expression and pathway enrichment of POLE2: Gene co-expression analysis is a powerful approach to uncover the biological functions and pathways associated with a target gene—in this case, POLE2 [17]. In the pan-cancer study, co-expression networks were constructed using both STRING and GEPIA2 platforms, identifying several genes with strong expression correlations to POLE2, such as DLGAP5, HELLS, MCM6, and TIMELESS. These genes are known to be involved in DNA replication, chromosomal dynamics, and mitotic control, suggesting that POLE2 operates within a tightly regulated proliferative network.
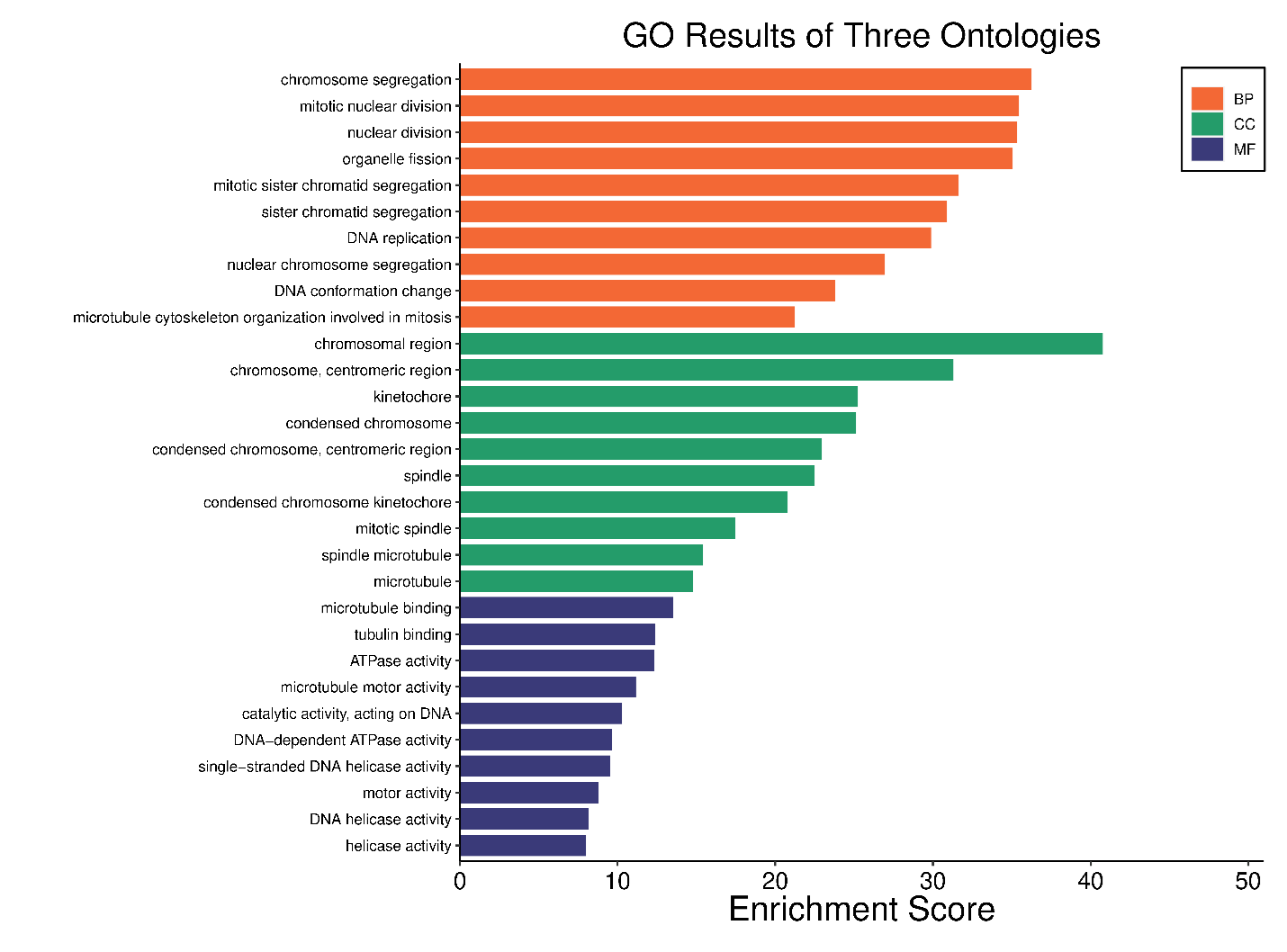
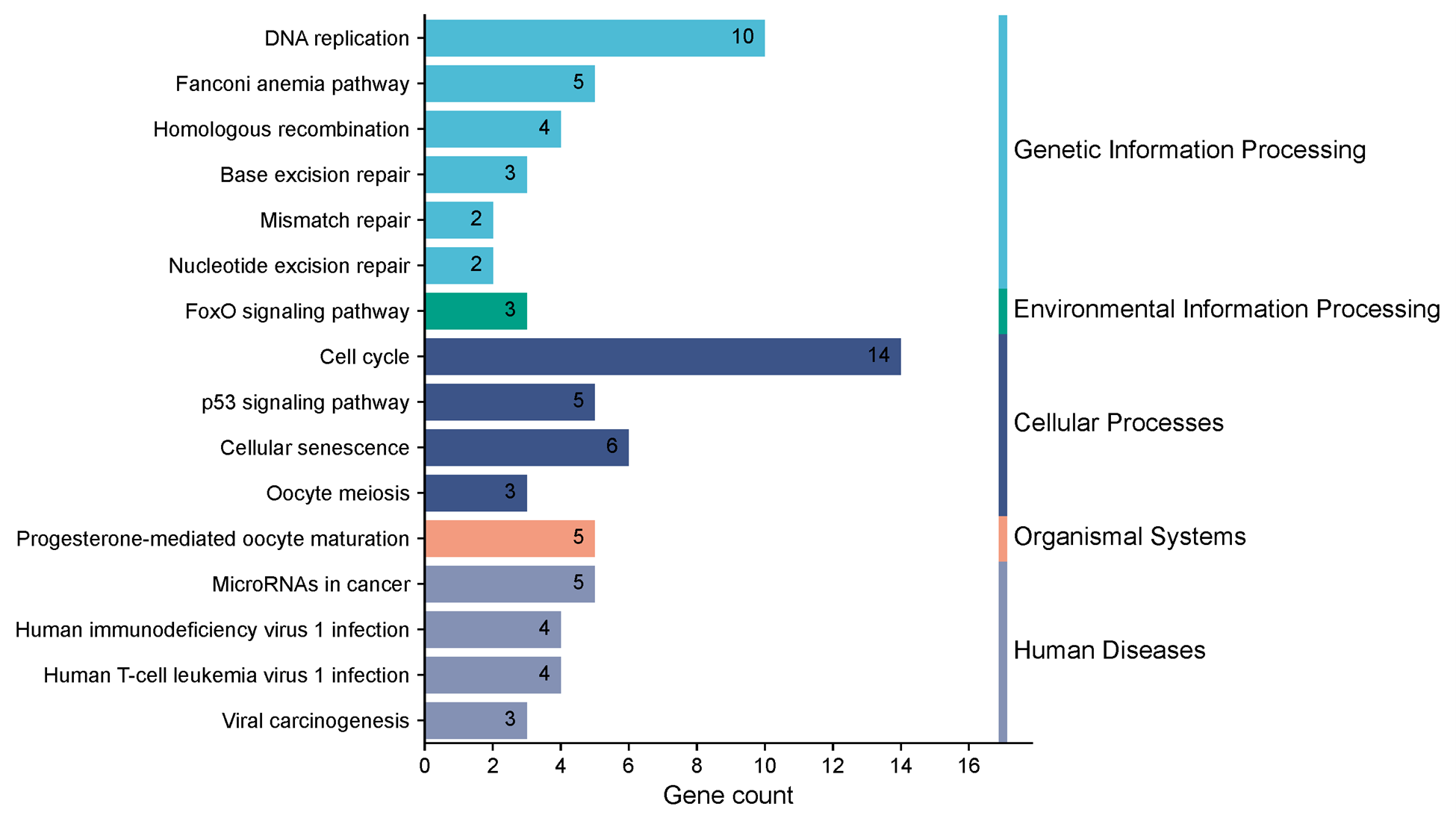
Pathway enrichment utilized Gene Ontology (GO) and KEGG analysis. Gene Ontology (GO) states that POLE2 mainly participates in mitosis, organelle fission, chromosomal segregation, mitotic nuclear division, DNA replication, and structural modifications in tumour cells in addition to organizing the microtubule cytoskeleton (Figure 3). Furthermore, the KEGG pathway provided important new insights into the role of POLE2 and its co-expressed genes in cellular degeneration, homologous recombination, base excision repair, mismatch repair, and the p53 signaling pathway, among other processes (Figure 4). These pathways are critical for maintaining genomic integrity, and their dysregulation can drive cancer progression. These pathways are often dysregulated in cancer and are critical for DNA damage sensing and repair. The connection between POLE2 and these pathways emphasizes its potential function as a guardian of genome stability—a role often exploited by cancer cells for unchecked proliferation.
Figure 3. Gene Ontology (GO) enrichment analysis of POLE2-associated genes across pan-cancer datasets. The bar plot illustrates the top enriched GO terms classified into three categories: Biological Process (BP, orange), Cellular Component (CC, green), and Molecular Function (MF, blue).
Figure 4. KEGG (Kyoto encyclopedia of genes and genomes) pathway enrichment of POLE2-associated genes in pan-cancer.
In summary, enrichment analyses firmly position POLE2 as a key player in fundamental oncogenic processes such as DNA replication, chromosomal segregation, and genome repair. Its strong association with cell cycle regulation and DNA repair pathways further supports its candidacy as a therapeutic target, particularly in cancers characterized by replication stress or genomic instability.
POLE2’s Role in cell proliferation and genomic instability: POLE2 plays a crucial role in cell proliferation through its function in the DNA polymerase epsilon complex, which ensures accurate DNA replication during the S phase of the cell cycle [1]. Aberrant expression of POLE2 has been linked to increased tumour cell division and poor clinical outcomes in multiple cancers. Additionally, its association with high tumour mutational burden (TMB) and microsatellite instability (MSI) in several tumour types suggests that POLE2 dysregulation may contribute to genomic instability—an established hallmark of cancer [18].
Our study found a strong positive correlation between POLE2 expression and TMB in 14 different tumour types, including BLCA, CHOL, COAD, KIRC, KICH, LAML, LGG, LIHC, LUAD, PRAD, PCPG, SKCM, STAD, and UCEC. TMB reflects the number of somatic mutations per megabase in tumor DNA, and higher TMB is typically associated with increased neoantigen load, making tumors more visible to the immune system. The positive relationship between POLE2 and TMB suggests that POLE2 may be linked to increased mutational processes, potentially through its role in DNA replication and repair. This supports the hypothesis that dysregulated POLE2 may contribute to replication stress and error-prone repair mechanisms that increase mutational load.
Similarly, POLE2 expression showed a significant positive correlation with MSI in 9 cancers, including COAD, KIRC, LUAD, OV, STAD, SARC, THCA, UCEC, and UCS, and a negative correlation in DLBC and TGCT. MSI arises from defects in the DNA mismatch repair (MMR) system, leading to widespread insertion/deletion mutations in microsatellites. The link between high POLE2 expression and MSI in certain cancers implies that POLE2 may interact with or reflect underlying MMR deficiencies. Given POLE2's involvement in DNA replication, overexpression may exacerbate errors in microsatellite regions when the MMR pathway is compromised.
These characteristics highlight POLE2's potential involvement in both tumour progression and the development of therapy resistance.
Supporting Evidence from Other Studies
Several other studies have demonstrated that POLE2 is involved in tumorigenesis. Pol ε is a multi-subunit complex composed of four subunits: the catalytic subunit encoded by the POLE gene, which possesses both polymerase and 3′–5′ exonuclease proofreading activities, and three accessory subunits, including POLE2. The exonuclease activity is crucial for correcting DNA synthesis errors, thereby preventing mutations and maintaining genome stability. The proofreading function of Pol ε is vital for high-fidelity DNA replication. Loss or impairment of this activity leads to increased replication errors, contributing to genomic instability—a hallmark of cancer [1], [19]. Mutations in the exonuclease domain of POLE have been identified in approximately 3% of colorectal and 7% of endometrial cancers. These mutations are associated with a hypermutator phenotype characterized by a high rate of single nucleotide substitutions (15–500 per megabase) while maintaining microsatellite stability. Such mutations have also been observed, albeit at lower frequencies, in other tumour types, suggesting a broader role in tumorigenesis [1]. MSI-high tumors exhibited significantly higher tumour mutational burden (TMB) than POLE-mutated tumors. They also harbored more insertion/deletion (indel) mutations, while POLE-mutated tumors showed predominantly single nucleotide variants. Some MSI-high tumors with concurrent POLE mutations of unknown significance also exhibited ultramutated profiles, suggesting potential combined pathogenic effects [20].
Conclusion
POLE2 is integral to cancer prognosis and immune modulation due to its role in DNA replication and genomic stability maintenance. A comprehensive pan-cancer analysis of POLE2 has demonstrated that increased expression of POLE2 is linked to unfavorable survival outcomes across various cancers, underscoring its prognostic significance. Furthermore, POLE2 exhibits a positive correlation with tumour mutational burden (TMB) and microsatellite instability (MSI), both of which are critical indicators of tumour immunogenicity. It also shows significant associations with immune checkpoint genes and immune cell infiltration, indicating its potential influence on the tumour immune microenvironment. These findings suggest that POLE2 may facilitate immune evasion and serve as a predictive biomarker for immunotherapy response. Although the pan-cancer analysis establishes POLE2 as a potential prognostic and immunological biomarker across diverse cancers, the specific molecular mechanisms by which POLE2 affects tumour progression and immune regulation remain to be elucidated. The observed associations with TMB, MSI, and immune checkpoint expression underscore POLE2's possible involvement in immune evasion and genomic instability. However, these bioinformatics-based findings require validation through mechanistic studies, such as in vitro and in vivo functional assays.
Competing Interests
The authors claim to have no conflicting interests.
Funding
No funding was given to the authors for this research.
Author Contributions
All authors were involved in the conception, design, data analysis, and drafting of the manuscript. All authors have reviewed and approved the final manuscript.
References
2. Zhu Y, Chen G, Song Y, Chen Z, Chen X. POLE2 knockdown reduce tumorigenesis in esophageal squamous cells. Cancer Cell Int. 2020 Aug 11;20:388.
3. Strauss JD, Pursell ZF. Replication DNA polymerases, genome instability and cancer therapies. NAR Cancer. 2023 Jun 28;5(3):zcad033.
4. Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016 Nov;49(11):607–11.
5. Stanfill AG, Cao X. Enhancing Research Through the Use of the Genotype-Tissue Expression (GTEx) Database. Biol Res Nurs. 2021 Jul;23(3):533–40.
6. Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022 Mar;25:18–27.
7. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020 Jul 2;48(W1):W509¬–W514.
8. Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015 Apr;25(4):198–213.
9. Liu Y, Zhou H, Zheng J, Zeng X, Yu W, Liu W, et al. Identification of Immune-Related Prognostic Biomarkers Based on the Tumor Microenvironment in 20 Malignant Tumor Types With Poor Prognosis. Front Oncol. 2020 Jul 31;10:1008.
10. Zeng Z, Li J, Zhang J, Li Y, Liu X, Chen J, et al. Immune and stromal scoring system associated with tumor microenvironment and prognosis: a gene-based multi-cancer analysis. J Transl Med. 2021 Aug 3;19(1):330.
11. Seager RJ, Hajal C, Spill F, Kamm RD, Zaman MH. Dynamic interplay between tumour, stroma and immune system can drive or prevent tumour progression. Converg Sci Phys Oncol. 2017;3:034002.
12. Chen X, Du Y, Lin X, Qian Y, Zhou T, Huang Z. CD4+CD25+ regulatory T cells in tumor immunity. Int Immunopharmacol. 2016 May;34:244–9.
13. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015 Apr 3;348(6230):56–61.
14. Viitanen TM, Sukumaran P, Löf C, Törnquist K. Functional coupling of TRPC2 cation channels and the calcium-activated anion channels in rat thyroid cells: implications for iodide homeostasis. J Cell Physiol. 2013 Apr;228(4):814–23.
15. Farber DL, Yudanin NA, Restifo NP. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol. 2014 Jan;14(1):24–35.
16. Baumjohann D, Brossart P. T follicular helper cells: linking cancer immunotherapy and immune-related adverse events. J Immunother Cancer. 2021 Jun;9(6):e002588.
17. Zogopoulos VL, Saxami G, Malatras A, Papadopoulos K, Tsotra I, Iconomidou VA, et al. Approaches in Gene Coexpression Analysis in Eukaryotes. Biology (Basel). 2022 Jul 6;11(7):1019.
18. Ahammad K, Jihan A, Akter S, Rahman A. Systematic pan-cancer analysis reveals the distinct role of POLE2 expression in cancer prognosis and its interaction with the tumour microenvironment. Next Research. 2025 May 7:100391.
19. Xing X, Jin N, Wang J. Polymerase Epsilon-Associated Ultramutagenesis in Cancer. Cancers (Basel). 2022 Mar 12;14(6):1467.
20. Hwang HS, Kim D, Choi J. Distinct mutational profile and immune microenvironment in microsatellite-unstable and POLE-mutated tumors. J Immunother Cancer. 2021 Oct;9(10):e002797.