Introduction
The field of mRNA modifications has rapidly developed over the last years, outlining a new realm of gene expression regulation that appears to play a major role in health and disease states including cancer. Considering the information regarding chemical modifications of DNA and proteins, decades of research proved beyond any doubt that chemical modifications affect chromatin structure as well as enzymatic activity. So, why did the field of RNA modifications and more specifically, mRNA modifications, lagged behind? The answer rests in the combination of technical difficulties to globally map these modifications and the low abundance of each mRNA transcript in total or even purified messenger RNA samples. The major breakthrough emerged in 2012 with the first approach to map N6-methyladenosine (m6A) in a transcriptome-wide manner [1,2], and the discovery of dedicated RNA binding proteins that exclusively recognize m6A-methylated sequences [1]. These proteins, later termed readers (see below), exert the roles of m6A in regulation of gene expression. In addition, m6A demethylases, termed erasers, grant this modification a dynamic nature, making it suitable for sensing changing conditions and adjusting the expression of genes accordingly [3,4]. Thus, m6A distribution and abundance together with its dynamics provide cells with a means to regulate sets of transcripts, enabling switching between transcription programs that are needed for transitions between cell states. For example, during early mouse embryonic stem cells differentiation, m6A promotes rapid degradation of mRNAs associated with pluripotency genes to safeguard linage priming and differentiation [5].
The discovery that mRNA m6A is essential for resolution of pluripotency raised the possibility that it may also play a role in cancer, or more specifically, changes in m6A mRNA methylation may lead to tumor formation and progression. In fact, global mapping of m6A in normal and cancer samples revealed differential methylation patterns of cancer-associated transcripts, reflecting dysregulation of its associated proteins and proving that m6A plays a critical role in almost every type of cancer [6–9].
In this mini-review, we outline the principles of m6A regulation, explain how imbalanced expression of m6A-associated proteins (also called m6A regulators) contributes to tumor development and discuss the harnessing of novel inhibitors of m6A-associated proteins to fight cancer. It is important to note that although m6A is present in mRNA of all eukaryotes, focusing on its part in cancer, we will discuss here only m6A data of human and mouse.
m6A Metabolism
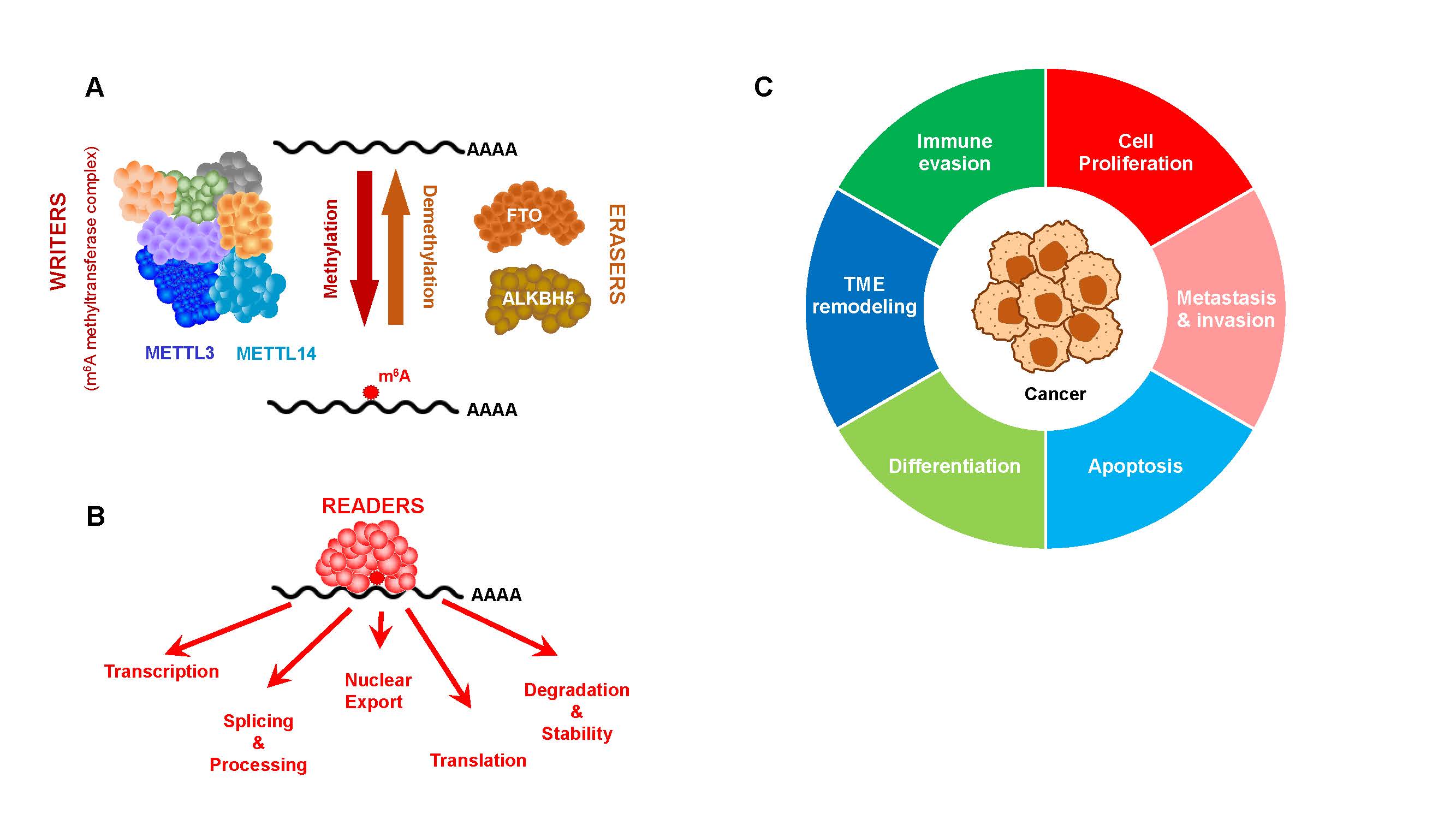
The notion that m6A is highly prevalent in mRNA underlined the need for its global mapping as a first step to unravel its roles. Indeed, transcriptome-wide mapping of mRNA m6A provided much more than just a collection of sites. The non-random site distribution of m6A was the first step in understanding its biological functions [1,2]. Further insights into the biology of m6A came from the identification of m6A-associated proteins: writers, erasers and readers (Figures 1A and 1B). The main writer complex of mRNA m6A includes METTL3, METTL14, WTAP, RBM15/15B, ZCH3H13, VIRMA and HAKAI, which together make the methyltransferase complex (MTC) [10]. The core subunit of MTC consists of a heterodimer of METTL3 and METTL14 (the catalytic and allosteric activator components, respectively). Other components are responsible for anchoring and stabilizing the interaction with target transcripts [10] (Figure 1A).
Figure 1. A. mRNA m6A is installed by writers that methylate mRNA transcripts. Demethylation is carried out by erasers, together granting m6A a dynamic nature. B. Reader proteins bind methylated transcripts to exert the regulatory roles of m6A, affecting transcription, splicing and processing, nuclear export, translation and stability/decay. In cancer, the most studied reader proteins belong to the YTH domain containing protein family (YTHDF1-3 and YTHDC1) and inulin-like growth factor 2 mRNA-binding proteins (IGF2BP1-3). C. m6A is implicated in carcinogenesis through its effects on the indicated processes.
Demethylation of mRNA m6A is carried out by two Fe2+- and 2-oxoglutarate (2OG)-dependent oxygenases, FTO [3] and ALKBH5 [4] (Figure 1A). These erasers function in the nucleus and the cytoplasm, conferring m6A its dynamic nature and enabling a rapid response to changing cellular and environmental cues.
Of particular significance are m6A reader proteins, which are RNA binding proteins that recognize methylated RNA sequences in a specific context and exert their regulatory roles [11,12]. An in-depth discussion of m6A readers and their multiple roles is beyond the scope of this mini-review and can be appreciated in dedicated reviews [13–16]. Yet, it is important to emphasize that m6A reader proteins affect multiple aspects of the mRNA life cycle including splicing, nuclear export, transcript decay and translation. Two major families of m6A reader proteins, the YTH domain containing family (YTHDF1-3 and YTHDC1-2, [17]) and the insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1-3 [18]), have been extensively implicated in cancer.
m6A Regulation of Signaling Pathways and Feedback Loops
Insights into regulatory mechanisms that involve m6A reveal its envelopment in activation of signaling pathways and in feedback loops. As m6A is involved in almost every aspect of gene expression, it is expected to affect multiple signaling cascades. The first indication for the implication of m6A in signaling emerged in 2012, showing that decreased METT3 levels modulated the p53 signaling pathway, through differential methylation, splicing and expression of 23 of its pathway members, including reduced levels of MDMX (also named MDM4), a negative regulator of p53 signaling cascade [1].
Since then, the role of m6A in regulation of additional signaling pathways was extensively studied, mostly of pathways that drive cancer (see more information in the next section). Research exposed how m6A rewires gene expression programs that are central to cancer, revealing that additional major oncogenic signal transduction pathways, such as PI3K/AKT, TGFβ, ERK, RAS/MAPK and the canonical Wnt pathway, are regulated by m6A. In such cases, m6A differentially methylates central proteins in these cascades during activation (as discussed in multiple reviews such as [19] and [20]). It is now well established that dysregulation of m6A leads to imbalanced activation of signaling pathways, which impacts cell proliferation, metastasis, and response/resistance to treatments (reviewed in publication such as [21,22]). For example, upregulation of METTL3 in prostate cancer [23] and hepatoblastoma [24], enhances the expression of b-catenin and LEF1 transcripts (both m6A-methylated), stabilizing b-catenin to activate the Wnt signaling pathway, stimulating cell proliferation and inhibiting differentiation [23,24].
Beyond direct modulation of signaling cascades, m6A also participates in intricate feedback mechanisms that link gene expression to chromatin state. Feedback loops involving m6A tightly regulate gene expression by bridging transcription dynamics with methylation and translation efficiency. Methylated transcripts recruit m6A readers that alter the chromatin state and transcriptional availability of specific regions in the genomic DNA. YTHDC1 binds methylated transcripts contributing to the formation of heterochromatin that restrain the activity of transposable elements in mouse embryonic stem cells [25]. In addition, m6A methylation of chromosome-associated regulatory RNAs (carRNAs), which physically interact with chromatin, decrease their stability and lead to a more open chromatin and active transcription [26].
Evidence for the existence of cross-talk between RNA and histone modifications uncovered the involvement of m6A in epigenetic regulation through m6A deposition on transcripts encoding histone modifying proteins. For instance, m6A destabilizes the histone acetyltransferase P300/CBP, mRNAs in neural stem cells. P300/CBP increases the prevalence of H3K27ac and H3K9ac, dictating a more relaxed accessible chromatin conformation, which facilitates transcription activation [27]. Moreover, m6A regulates H3K9me2 (associated with transcription repression) through YTHDC1. The latter recruits the H3K9me2 demethylase, KDM3B, coupling m6A with heterochromatin state [28]. Such intertwined feedback circuits are critical for cell homeostasis, development and some disease states [29].
Expanding this regulatory landscape, a large body of evidence demonstrates that mRNA modifications beyond m6A also regulate gene expression, with indications for crosstalk between different mRNA modifications (reviewed in [30]). In fact, the mRNA modification N1-methyladenosine (m1A) accelerates degradation of transcripts co-modified with m6A [31]. Another study predicted the co-occurrence of m6A and 5-methylcytosine (m5C), based on a novel tool for analyzing direct RNA sequencing data (CHEUI, CH3 Estimation Using Ionic current). CHEUI analysis revealed non-random profiles of the two modifications, co-occurring on the same transcripts. This finding supports an interplay between these two mRNA marks [32].
Epigenetic mechanisms, including DNA methylation and histone modifications, demonstrate an interplay with mRNA modifications as well. m?A and m5C were both associated with influencing chromatin state [33,34]. Chemical modifications of proteins other than histones are also at play. Interesting discovery shows that m6A levels are regulated by symmetric dimethylarginine modification of METTL14, demonstrating a novel interplay between post-translational protein modifications of m6A writers and m6A levels [35]. Together, these findings sketch a convoluted network of regulation that is central in health and diseases, of which only the tip of the iceberg seems to be exposed.
The Roles of m6A in Cancer
Cancer cells often exhibit altered expression of subsets of genes that act in concert to maintain the hallmarks of cancer, including sustained proliferation, resistance to cell death, replicative immortality, angiogenesis, invasiveness, metabolic reprogramming and immune evasion (Figure 1C) [36]. It is therefore not surprising that m6A machinery is disrupted in many cancers [37]. In fact, alterations in m6A levels have been shown to affect almost every cancer hallmarks. Interestingly, dysregulation of m6A-associated proteins may result in opposite effects in different types of cancer and even within the same cancer type, in a context specific manner (see examples in [38]). Here, we focus on two key major hallmarks of cancer – chronic proliferation and immune evasion. The role of m6A in the regulation of other cancer hallmarks is discussed elsewhere [39–42].
Sustained proliferation can be acquired by excessive production of growth factors, overexpression of growth factor receptors and constitutive activation of signaling pathways downstream of growth receptors. Accumulated data revealed that dysregulation of m6A is implicated in all these mechanisms. For instance, increased m6A modification promotes the translation of epidermal growth factor receptor (EGFR), a tyrosine kinase receptor, in a wide range of tumors such as lung adenocarcinoma [14], hepatocellular carcinoma [43] and melanoma [44], activating downstream signaling pathways and acting as an oncogene. In addition, increased levels of m6A methylation drives overexpression of other oncogenic tyrosine kinase receptors, such as c-Met, in different cancer types including breast [45], gastric [46] and melanoma [45]. Alternatively, dysregulated deposition and demethylation of m6A can result in overexpression of transcription factors (TFs) that drive proliferation. One prominent example is that of the master transcription factor, MYC [47]. On the whole, unbalanced expression of TFs skews the expression of their targets, resulting in changes of gene expression across multiple genes, simultaneously. In fact, increased expression of METTL3 [48], METTL14 [49] and WTAP [50] as well as the eraser, FTO [51], modulates m6A levels to enhance MYC mRNA stability and translation in acute myeloid leukemia (AML). Furthermore, overexpression of IGF2BPs, is evident in numerous types of cancers: MYC and its homologue MYCN are regulated by IGF2BP1 and IGF2BP3, which stabilize methylated target transcripts in cholangiocarcinoma [52], gastric cancer (where it inhibits proliferation) [53], neuroblastoma [54,55] and nasopharyngeal carcinoma [56]. Of note, YTHDF2, which destabilizes mRNA transcripts, has been shown to exert an opposite effect on MYC in glioblastoma [46]. Importantly, while overexpression of MYC enhances the tumorigenic phenotype, reducing its levels contributes to the ability of cells to escape chemotherapy [57–59]. Such convoluted and opposite effects of m6A highlight its dual role resulting in either pro or anti-tumorigenic effects depending on the specific cellular and molecular context.
In order to survive, cancer cells employ various mechanisms to evade the multiple arms of the immune systems including the downregulation of antigen presentation, suppression of the major histocompatibility complex (MHC) I, upregulation immune checkpoints, such as PD-L1, PD-L2, and B7-H3, recruitment of immunosuppressive cells to the tumor microenvironment (TME), such as T regulatory cells and M2 macrophages.
m6A has been to shown to regulate the expression of the surface expression of MHC-I. For example, in melanoma cells, YTHDF1 was shown to increase the expression of lysosomal genes, enhancing MHC-I degradation, driving immune evasion, and resistance to immunotherapy, promoting tumor survival [60]. Furthermore, YTHDF1 silencing promoted cell cycle arrest of pancreatic and breast cancer cells, leading to inhibition of lysosomal protein translation and upregulation of MHC-I expression [61].
Dysregulation of m6A writers and erasers has also been shown to remodel the TME compartment of multiple tumor types. For instance, imbalanced m6A patterns, due to changes in METTL3/METTL14 expression levels, impacts T cells homeostasis, activation, differentiation and the release of inflammatory cytokines through m6A-mediated degradation of mRNAs [62,63]. The balance between different subpopulations of T cells in the TME is critical for anti-tumor immunity. A recent pan-cancer analysis revealed that m6A methylation by METTL3 mediates T cell exhaustion [64], enabling cancer cells to evade the cytotoxic effect of T cells. Moreover, deletion of METTL3 in natural killer (NK) cells inhibited NK cells infiltration in melanoma models and rendered them hyposensitive to IL-15 [65]. Finally, inhibition of METTL3 in melanoma models has been shown to polarize tumor associated macrophages towards a more immunosuppressive M2 phenotype [66].
In addition, several recent studies suggest that the m6A machinery regulates the expression of PD-L1 and the secretion of pro- and anti-inflammatory signals by cancer cells. However, m6A writers exhibit a complex role in cancer immunity. Knockout of METTL3 and METTL14 enhanced the anti-PD-L1 therapy efficacy and stimulated the secretion of pro-inflammatory cytokines (including IFN-γ, CXCL9 and CXCL10) [67]. In another study, METTL3 was found to promote PD-L1 expression of via IGF2BP3-dependent mechanisms in breast cancer [68]. In addition, inhibition of FTO downregulates PD-L1 and LILRB4 in AML cells and sensitizes them to T cell cytotoxicity [69]. Furthermore, inhibition of ALKBH5 potentiates the response to immune checkpoint blockade in melanoma and colon cancer by modulating the composition of the TME [70] and by increasing PD-L1 expression in intrahepatic cholangiocarcinoma [71].
In conclusion, the normal phenotype is a result of a highly concerted equilibrium of m6A writers, erasers and readers. Any deviation from this balance is prone to lead to dysregulated cell states resulting in rapid proliferation, halted differentiation and escape from apoptosis.
Targeting m6A Machinery to Fight Cancer
Targeting the m6A machinery has emerged as a promising therapeutic strategy [72]. Recent advances in our understanding of m6A biology has prompted the development of specific small molecule inhibitors of m6A regulators. Screening for such inhibitors was successful due to the improved and more accurate structural data of m6A regulators, defining targetable domains to impair their actions. Importantly, most such inhibitors are still in their discovery phase, but a few have entered clinical trials. The field of m6A inhibitors is rapidly developing [73,74]. In this mini-review, we focus on several examples of inhibitors, their mechanism of action and pharmacokinetics:
Inhibiting m6A writers, such as METTL3 and METTL14 leads to reduced mRNA m6A levels, which affect the expression of many genes, including oncogenes. The METTL3 inhibitor, STM2457, potently blocks its enzymatic activity at low nanomolar range (IC50 of 16.9 nM in AML cells [75]) by competitively binding the S-adenosylmethionine (SAM, the methyl donor) pocket, with high affinity (Kd=1.4 mM) [75]. Further improvement of this inhibitor generated STM3006, which exhibits a greater cellular potency, having higher efficiency and affinity in AML cells (Kd=55 pM, IC50=5 nM) [76]. Both inhibitors showed in vitro and in vivo efficacy in AML, resulting in apoptosis, increased secretion of IFN-alpha (-a) [76] and triggering activation of IFN-a response [76,77]. The first METTL3 inhibitor that entered clinical trials, STC-15, an improved derivative of STM2457 [78], is now in Phase I for patients with refractory AML (NCT05584111) [79]. Similar to STM2457 and STM3006, STC-15 also activates the IFN-a signaling response, remodeling the TME towards pro-inflammatory state and downregulates the anti-apoptotic protein BCL-2 [73]. Phase I results showed that STC-15 is well-tolerated across a range of doses in AML patients which supports the development of subsequent phase trials for STC-15 (http://www.stormtherapeutics.com/news/press-releases/storm-therapeutics-announces-first-patient-dosed-in-clinical-collaboration-to-evaluate-stc-15-in-combination-with-loqtorzi/). Inhibition of METTL3 results in reduced cell proliferation, increased apoptosis and remodeling of the TME in additional cancer types including colorectal cancer, ovarian cancer, thyroid cancer prostate cancer, non-small cell lung cancer and myelodysplastic neoplasms [76,80–85].
Inhibition of m6A erasers, FTO or ALKBH5, results in increased mRNA m6A levels, which has broad consequences [86]. FTO inhibition by several tool compounds including FB23, FB23-2 and R-2HG showed increased killing of AML cells. Clinical FTO inhibitors, CS1 (bisantrene) and CS2 (brequinar), exhibit high inhibitory potential, with IC50 values in the nanomolar range [69], resulting in suppression of AML stem cells and decreased expression of the immune checkpoint gene, LILRB4 [51,69]. Early phase clinical trials using bisatrene for the treatment of AML have shown both safety and efficacy (NCT04989335) [87]. A Phase II study of bisantrene in relapsed/refractory AML (NCT03820908) reported a 40% overall response rate with low toxicity [87]. Of note, inhibitors of ALKBH5 and m6A readers are still in their infancy. FTO inhibition by CS1 and CS2 induced increased methylation on MYC, promoting its transcript degradation and reducing its expression, suppressing the MYC targets V1 and V2 pathways, and exerting anti-cancer effects [69,88,89]. Moreover, CS1 and CS2 suppress PI3K/AKT pathway activation caused by FTO overexpression in several cancer cell lines [90–92]. Development of clinical specific inhibitors allows for their incorporation into combination regimens for cancer treatment. Table 1 lists examples of successive m6A-mediated approaches for combining m6A inhibitors with other cancer therapy modalities.
|
Target |
m6A inhibitor |
Treatment |
Outcome |
Cancer type |
ref |
|
METTL3 |
STM2457 STM3006 STC-15 |
Chemotherapy (PTX, CBP, venetoclax, anlotinib, dox |
|
NSCLC ESCC OSCC AML NBL SCLC BC OC |
[98–104] |
|
Immunotherapy (anti-PD1; anti-VISTA; anti-BET (HJP-178 and OTX-015)) |
|
Melanoma OC GA NSCLC CRC BLC AML |
[76,95,105–9] |
||
|
FTO |
CS1 CS2 FB23/FB23-2 Rhein
|
Chemotherapy (PTX; 5-FU; |
|
AML NBL CRC GIC Melanoma
|
[69,110–113] |
|
Immunotherapy (anti-PD1) |
|
AML HCC Melanoma |
[114–116] |
||
|
PTX: Paclitaxel; CBP: Carboplatin; Dox: Doxorubicin; Bromodomain and Extra-Terminal domain proteins; VISTA: V-domain Immunoglobulin Suppressor of T cell Activation; 5-FU: 5-fluorouracil; NSCLC: Non-Small Cell Lung Cancer; ESCC: Esophageal Squamous Cell Carcinoma; OSCC: Oral Squamous Cell Carcinoma; BC: Breast Cancer; NBL: Neuroblastoma; OC: Ovarian Cancer; GA: Gastric Cancer; BLC: Bladder Cancer; HCC: Hepatocellular Carcinoma |
|||||
The most widely used therapy modality is chemotherapy and several studies have shown that inhibition of METTL3 or FTO can sensitize cancer cells to chemotherapy. For instance, inhibition of METTL3 enhanced the cytotoxic effects of cisplatin in ovarian cancer [93] and doxorubicin in AML [94]. Furthermore, immunotherapy, which has revolutionized the field of cancer, can also be combined with m6A inhibitors. For example, inhibition of METTL3 in AML and melanoma has been shown to upregulate PD-L1, enhancing the efficacy of immune checkpoint inhibition [77]. Furthermore, inhibition of FTO results in downregulation of the immune checkpoint gene LILRB4 and enhances cytotoxicity of T cells [69]. Modulation of m6A regulators has also been shown to affect the immune landscape of several tumors. For instance, inhibition of METTL3 suppressed the function of regulatory T cells in melanoma and colorectal adenocarcinoma [95]. Importantly, while inhibition of METTL3 may decrease the viability of cancer cells and enhance T cell cytotoxicity, it has also been shown to promote polarization from M1 macrophages towards M2 phenotype [66], thus representing a double edge sword. In this respect, studies in AML, using METTL3 [79] and FTO inhibitors [87] demonstrate two complementary but mechanistically opposed therapeutic strategies, achieving success through inverse manipulation of the same epitranscriptomic mark.
Future Prospective
The development of m6A inhibitors marks a significant step toward translating our accumulating knowledge regarding this prevalent mRNA modification into new regimes to fight cancer. With the emergence of clinical trials testing the efficacy of m6A inhibitors in various cancer types, we anticipate a growing number of additional clinical trials combining this strategy with chemotherapy, radiation, immune checkpoint inhibitors, and chimeric antigen receptor (CAR) T cells.
Despite the promising advances of m6A inhibitors, several obstacles remain to be addressed prior to development and clinical application of m6A-targeted therapies. One of the major challenges is the potential development of therapy resistance to these inhibitors. Since the use of this new class of drugs is in its infancy, little is known about mechanisms of therapy resistance warranting future research. Moreover, depletion of m6A writers and erasers can have pervasive consequences [5,96], making it imperative to determine the safe and effective window of each drug. In addition, as m6A modifications are ubiquitous in normal tissues, side effects are expected to occur in a dose dependent manner. The toxicity profile of the m6A inhibitors is still being investigated, however, the FTO inhibitor CS1 (bisantrene) was well tolerated in a phase II clinical trial at our institution [87]. Results from ongoing clinical trials will shed more light on the toxicity of m6A inhibitors.
Finally, it is important to remember that the effect of a specific m6A inhibitor results from the disruption of m6A methylation on multiple transcripts in parallel. Due to their lack of specificity, such drugs are anticipated to have a wide “off-target” effect. Recent advancements in nucleic acid editing supports the development of targeted RNA editing technology using the inactive Cas13 (dCas13) enzyme coupled to FTO [97], enabling the m6A manipulation of a single transcript. While this technology is still restricted for basic research purposes, targeting specific m6A sites in oncogenes or tumor suppressors using CRISPR-dCas13 may emerge as a novel and effective therapeutic strategy.
References
2. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012 Jun 22;149(7):1635-46.
3. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011 Oct 16;7(12):885-87.
4. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013 Jan 10;49(1):18-29.
5. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015 Feb 27;347(6225):1002–6
6. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019 Dec 4;18(1):176.
7. Hu H, Li Z, Xie X, Liao Q, Hu Y, Gong C, et al. Insights into the role of RNA m6A modification in the metabolic process and related diseases. Genes Dis. 2023 Jul 4;11(4):101011.
8. Bhattarai PY, Kim G, Bhandari D, Shrestha P, Choi HS. Regulation of m6A Methylome in Cancer: Mechanisms, Implications, and Therapeutic Strategies. Cells. 2023 Dec 28;13(1):66.
9. Liu J, Huang H, Zhang M, Qing G, Liu H. Intertwined regulation between RNA m6A modification and cancer metabolism. Cell Insight. 2022 Dec 5;2(1):100075.
10. Garcias Morales D, Reyes JL. A birds'-eye view of the activity and specificity of the mRNA m6A methyltransferase complex. Wiley Interdiscip Rev RNA. 2021 Jan;12(1):e1618.
11. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016 Aug 25;7:12626.
12. Liu J, Gao M, Xu S, Chen Y, Wu K, Liu H, et al. YTHDF2/3 Are Required for Somatic Reprogramming through Different RNA Deadenylation Pathways. Cell Rep. 2020 Sep 8;32(10):108120.
13. Chen Y, Zhou Z, Chen Y, Chen D. Reading the m6A-encoded epitranscriptomic information in development and diseases. Cell Biosci. 2024 Sep 28;14(1):124.
14. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell. 2016 May 5;62(3):335-45.
15. Duan M, Liu H, Xu S, Yang Z, Zhang F, Wang G, et al. IGF2BPs as novel m6A readers: Diverse roles in regulating cancer cell biological functions, hypoxia adaptation, metabolism, and immunosuppressive tumor microenvironment. Genes Dis. 2023 Jul 20;11(2):890-920.
16. Zhao Y, Shi Y, Shen H, Xie W. m6A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020 Apr 10;13(1):35.
17. Liao S, Sun H, Xu C. YTH Domain: A Family of N6-methyladenosine (m6A) Readers. Genomics Proteomics Bioinformatics. 2018 Apr;16(2):99-107.
18. Cai Y, Wang Y, Mao B, You Q, Guo X. Targeting insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) for the treatment of cancer. Eur J Med Chem. 2024 Mar 15;268:116241.
19. Uddin MB, Wang Z, Yang C. The m6A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol Cancer. 2021 Apr 4;20(1):61.
20. Jang KH, Heras CR, Lee G. m6A in the Signal Transduction Network. Mol Cells. 2022 Jul 31;45(7):435-43.
21. Afrasiyab AS, Zheng P, Huang S. RNA m6A Modifications and Cancer. Int J Oncol Res. 2023;6:055.
22. Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021 Feb 21;6(1):74.
23. Ma XX, Cao ZG, Zhao SL. m6A methyltransferase METTL3 promotes the progression of prostate cancer via m6A-modified LEF1. Eur Rev Med Pharmacol Sci. 2020 Apr;24(7):3565-571.
24. Liu L, Wang J, Sun G, Wu Q, Ma J, Zhang X, et al. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer. 2019 Dec 23;18(1):188.
25. Xu W, Li J, He C, Wen J, Ma H, Rong B, et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. 2021 Mar;591(7849):317-21.
26. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020 Jan 31;367(6477):580–86.
27. Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018 Feb;21(2):195-206.
28. Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, et al. N6-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020 Sep;52(9):870-7.
29. Akhtar J, Lugoboni M, Junion G. m6A RNA modification in transcription regulation. Transcription. 2021 Oct;12(5):266-76.
30. Zhao Y, Chen Y, Jin M, Wang J. The crosstalk between m6A RNA methylation and other epigenetic regulators: a novel perspective in epigenetic remodeling. Theranostics. 2021 Mar 4;11(9):4549-566.
31. Boo SH, Ha H, Kim YK. m1A and m6A modifications function cooperatively to facilitate rapid mRNA degradation. Cell Rep. 2022 Sep 6;40(10):111317.
32. Acera Mateos P, J Sethi A, Ravindran A, Srivastava A, Woodward K, Mahmud S, et al. Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications. Nat Commun. 2024 May 9;15(1):3899.
33. Alagia, A. and M. Gullerova, Alagia A, Gullerova M. The Methylation Game: Epigenetic and Epitranscriptomic Dynamics of 5-Methylcytosine. Front Cell Dev Biol. 2022 Jun 3;10:915685.
34. Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature. 2019 Mar;567(7748):414-419.
35. Zhong Y, Zhang R, Lu L, Tan H, You Y, Mao Y, Yuan Y. Specific sDMA modifications on the RGG/RG motif of METTL14 regulate its function in AML. Cell Commun Signal. 2025 Mar 8;23(1):126.
36. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646-74.
37. Sarraf G, Chhabra R. Emerging role of mRNA methylation in regulating the hallmarks of cancer. Biochimie. 2023 Mar;206:61-72.
38. Moshitch-Moshkovitz S, Sevilla-Sharon M, Ashwal-Fluss R, Glick-Saar E, Rechavi G, Dominissini D. mRNA m6A detection. Nature Reviews Methods Primers. 2024;4(1):87.
39. Zhou Y, Cao P, Zhu Q. The regulatory role of m6A in cancer metastasis. Front Cell Dev Biol. 2025 Apr 28;13:1539678.
40. Liu L, Li H, Hu D, Wang Y, Shao W, Zhong J, et al. Insights into N6-methyladenosine and programmed cell death in cancer. Mol Cancer. 2022 Jan 28;21(1):32.
41. An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022 Jan 12;21(1):14.
42. Qin L, Zeng X, Qiu X, Chen X, Liu S. The role of N6-methyladenosine modification in tumor angiogenesis. Front Oncol. 2024 Dec 3;14:1467850.
43. Wang L, Yang Q, Zhou Q, Fang F, Lei K, Liu Z, et al. METTL3-m6A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023 Apr 10;559:216122.
44. Bhattarai PY, Kim G, Poudel M, Lim SC, Choi HS. METTL3 induces PLX4032 resistance in melanoma by promoting m6A-dependent EGFR translation. Cancer Lett. 2021 Dec 1;522:44-56.
45. Luo G, Xu W, Zhao Y, Jin S, Wang S, Liu Q, et al. RNA m6A methylation regulates uveal melanoma cell proliferation, migration, and invasion by targeting c-Met. J Cell Physiol. 2020 Oct;235(10):7107-19.
46. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021 Feb;11(2):480-99.
47. Zheng L, Li M, Wei J, Chen S, Xue C, Zhan Y, et al. The emerging roles of the interaction between m6A modification and c-Myc in driving tumorigenesis and development. J Cell Physiol. 2022 Jul;237(7):2758-69.
48. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017 Nov;23(11):1369-76.
49. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018 Feb 1;22(2):191-205.e9.
50. Naren D, Yan T, Gong Y, Huang J, Zhang D, Sang L, et al. High Wilms' tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m6A methylation of MYC mRNA. J Cancer Res Clin Oncol. 2021 Jan;147(1):33-47.
51. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell. 2018 Jan 11;172(1-2):90-105.e23.
52. Xiao P, Meng Q, Liu Q, Lang Q, Yin Z, Li G, et al. IGF2BP1-mediated N6-methyladenosine modification promotes intrahepatic cholangiocarcinoma progression. Cancer Lett. 2023 Mar 31;557:216075.
53. Ding N, Cao G, Wang Z, Xu S, Chen W. Tumor suppressive function of IGF2BP1 in gastric cancer through decreasing MYC. Cancer Sci. 2024 Feb;115(2):427-38.
54. Zhu K, Gao T, Wang Z, Zhang L, Tan K, Lv Z. RNA N6-methyladenosine reader IGF2BP3 interacts with MYCN and facilitates neuroblastoma cell proliferation. Cell Death Discov. 2023 May 8;9(1):151.
55. Hagemann S, Misiak D, Bell JL, Fuchs T, Lederer MI, Bley N, et al. IGF2BP1 induces neuroblastoma via a druggable feedforward loop with MYCN promoting 17q oncogene expression. Mol Cancer. 2023 May 29;22(1):88.
56. Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J, et al. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022 Feb 8;8(1):53.
57. Grossmann LD, Chen CH, Uzun Y, Thadi A, Wolpaw AJ, Louault K, et al. Identification and Characterization of Chemotherapy-Resistant High-Risk Neuroblastoma Persister Cells. Cancer Discov. 2024 Dec 2;14(12):2387-2406.
58. Ramesh-Kumar D, Guil S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. 2022 Nov;86(Pt 3):18-31.
59. Li D, Hu S, Ye J, Zhai C, Liu J, Wang Z, et al. The Emerging Role of IGF2BP2 in Cancer Therapy Resistance: From Molecular Mechanism to Future Potential. Int J Mol Sci. 2024 Nov 12;25(22):12150.
60. Lin W, Chen L, Zhang H, Qiu X, Huang Q, Wan F, et al. Tumor-intrinsic YTHDF1 drives immune evasion and resistance to immune checkpoint inhibitors via promoting MHC-I degradation. Nat Commun. 2023 Jan 17;14(1):265.
61. Liu H, Zhang X, Ding F, Pan J, Zhu H, Zhou Z, et al. YTHDF1-targeting nanoassembly reverses tumoral immune evasion through epigenetics and cell cycle modulation. J Control Release. 2025 May 10;381:113574.
62. Ito-Kureha T, Heissmeyer V. Critical functions of N6-adenosine methylation of mRNAs in T cells. Biochim Biophys Acta Mol Cell Res. 2023 Jan;1870(1):119380.
63. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017 Aug 17;548(7667):338-42.
64. Ji W, Fang Y, Chen L, Zheng Y, Pei Y, Mei C, et al. Pan-cancer characterization of m6A-mediated regulation of T cell exhaustion dynamics and clinical relevancies in human cancers. Mol Ther Nucleic Acids. 2025 Jan 25;36(1):102465.
65. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m6A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun. 2021 Sep 17;12(1):5522.
66. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun. 2021 Mar 2;12(1):1394.
67. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. m6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020 Oct 15;39(20):e104514.
68. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022 Feb 23;21(1):60.
69. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020 Jul 13;38(1):79-96.e11.
70. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H, et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):20159-0170.
71. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res. 2021 Sep 15;81(18):4778-93.
72. Pan J, Huang T, Deng Z, Zou C. Roles and therapeutic implications of m6A modification in cancer immunotherapy. Front Immunol. 2023 Mar 7;14:1132601.
73. Wu Z, Smith AR, Qian Z, Zheng G. Patent landscape of small molecule inhibitors of METTL3 (2020-present). Expert Opin Ther Pat. 2024 Dec 26:1-16.
74. Huang Y, Xia W, Dong Z, Yang CG. Chemical Inhibitors Targeting the Oncogenic m6A Modifying Proteins. Acc Chem Res. 2023 Nov 7;56(21):3010-22.
75. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021 May;593(7860):597-601.
76. Guirguis AA, Ofir-Rosenfeld Y, Knezevic K, Blackaby W, Hardick D, Chan YC, et al. Inhibition of METTL3 Results in a Cell-Intrinsic Interferon Response That Enhances Antitumor Immunity. Cancer Discov. 2023 Oct 5;13(10):2228-247.
77. Brichkina A, Suezov R, Huber M. Methyltransferase-like 3 (METTL3) inhibition potentiates anti-tumor immunity: a novel strategy for improving anti-PD1 therapy. Signal Transduct Target Ther. 2023 Dec 11;8(1):448.
78. Ofir-Rosenfeld Y, Vasiliauskaitė L, Saunders C, Sapetschnig A, Tsagkogeorga G, Albertella M, et al. STC-15, an oral small molecule inhibitor of the RNA methyltransferase METTL3, inhibits tumour growth through activation of anti-cancer immune responses associated with increased interferon signalling, and synergises with T cell checkpoint blockade. European Journal of Cancer. 2022 Oct 1;174:S123.
79. Moser JC, Papadopoulos KP, Rodon Ahnert J, Ofir-Rosenfeld Y, Holz JB, STC15-22101 Study Team. Phase 1 dose escalation and cohort expansion study evaluating safety, PK, PD and clinical activity of STC-15, a METTL-3 inhibitor, in patients with advanced malignancies. Journal of Clinical Oncology, 2024. 42(16_suppl): p. 2586.
80. Sun Y, Gong W, Zhang S. METTL3 promotes colorectal cancer progression through activating JAK1/STAT3 signaling pathway. Cell Death Dis. 2023 Nov 25;14(11):765.
81. Zhang H, Wang X, Chen J, Su R. Combating cancer stem cells: RNA m6A methylation and small-molecule drug discovery. Frontiers in Drug Discovery. 2024 Sep 30;4:1465222.
82. Zhou M, Zhang Y, Zhang Q, Tong Y. METTL14-mediated m6A modification upregulated SOCS3 expression alleviates thyroid cancer progression by regulating the JAK2/STAT3 pathway. Mol Cell Probes. 2024 Dec;78:101987.
83. Chen X, Wang M, Wang H, Yang J, Li X, Zhang R, et al. METTL3 inhibitor suppresses the progression of prostate cancer via IGFBP3/AKT pathway and synergizes with PARP inhibitor. Biomed Pharmacother. 2024 Oct;179:117366.
84. Xuan YF, Lu S, Ou YJ, Bao XB, Huan XJ, Song SS, et al. The combination of methionine adenosyltransferase 2A inhibitor and methyltransferase like 3 inhibitor promotes apoptosis of non-small cell lung cancer cells and produces synergistic anti-tumor activity. Biochem Biophys Res Commun. 2024 Jul 5;716:150011.
85. Jiang L, Zhang Y, Qian J, Zhou X, Ma L, Zhu S, et al. The m6A methyltransferase METTL14 promotes cell proliferation via SETBP1-mediated activation of PI3K-AKT signaling pathway in myelodysplastic neoplasms. Leukemia. 2024 Oct;38(10):2246-58.
86. Qiu L, Jing Q, Li Y, Han J. RNA modification: mechanisms and therapeutic targets. Mol Biomed. 2023 Aug 24;4(1):25.
87. Canaani J, Danylesko I, Shemtov N, Zlotnick M, Lozinsky K, Benjamini O, et al. A phase II study of bisantrene in patients with relapsed/refractory acute myeloid leukemia. Eur J Haematol. 2021 Feb;106(2):260-6.
88. Su R, Dong L, Li Y, Han L, Gao M, Wunderlich M, et al. Effective novel FTO inhibitors show potent anti-cancer efficacy and suppress drug resistance. Blood. 2019 Nov 13;134:233.
89. Zhang Z, Gao Q, Wang S. Kinase GSK3β functions as a suppressor in colorectal carcinoma through the FTO-mediated MZF1/c-Myc axis. J Cell Mol Med. 2021 Mar;25(5):2655–65.
90. Zhou LL, Xu H, Huang Y, Yang CG. Targeting the RNA demethylase FTO for cancer therapy. RSC Chem Biol. 2021 Jul 26;2(5):1352-69.
91. Liu Y, Wang R, Zhang L, Li J, Lou K, Shi B. The lipid metabolism gene FTO influences breast cancer cell energy metabolism via the PI3K/AKT signaling pathway. Oncol Lett. 2017 Jun;13(6):4685-90.
92. Phan T, Nguyen VH, Su R, Li Y, Qing Y, Qin H, et al. Targeting fat mass and obesity-associated protein mitigates human colorectal cancer growth in vitro and in a murine model. Front Oncol. 2023 Feb 17;13:1087644.
93. Xia Q, Zhong R, Zheng J, Zhou X, Zhao X, Wang S, et al. PRMT5-mediated methylation of METTL3 promotes cisplatin resistance in ovarian cancer by facilitating DNA repair mechanisms. Cell Rep. 2025 Apr 22;44(4):115484.
94. Li M, Ye J, Xia Y, Li M, Li G, Hu X, et al. METTL3 mediates chemoresistance by enhancing AML homing and engraftment via ITGA4. Leukemia. 2022 Nov;36(11):2586-95.
95. Wu K, Li S, Hong G, Dong H, Tang T, Liu H, et al. Targeting METTL3 as a checkpoint to enhance T cells for tumour immunotherapy. Clin Transl Med. 2024 Nov;14(11):e70089.
96. Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009 Jul;85(1):106-11.
97. Wilson C, Chen PJ, Miao Z, Liu DR. Programmable m6A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat Biotechnol. 2020 Dec;38(12):1431-40.
98. Zhang R, Chen P, Wang Y, Zeng Z, Yang H, Li M, et al. Targeting METTL3 enhances the chemosensitivity of non-small cell lung cancer cells by decreasing ABCC2 expression in an m6A-YTHDF1-dependent manner. Int J Biol Sci. 2024 Sep 3;20(12):4750-66.
99. Gao C, Yang H, Cheng J, He S, Yang Y, Xu L, et al. STM2457 impairs the proliferation of esophageal squamous cell carcinoma by activating DNA damage response through ATM-Chk2 axis. Med Oncol. 2025 Feb 22;42(3):82.
100. Jiao CQ, Hu C, Sun MH, Li Y, Wu C, Xu F, et al. Targeting METTL3 mitigates venetoclax resistance via proteasome-mediated modulation of MCL1 in acute myeloid leukemia. Cell Death Dis. 2025 Apr 1;16(1):233.
101. Liu L, Zhao T, Zheng S, Tang D, Han H, Yang C, et al. METTL3 inhibitor STM2457 impairs tumor progression and enhances sensitivity to anlotinib in OSCC. Oral Dis. 2024 Oct;30(7):4243-54.
102. Thombare K, Vaid R, Pucci P, Ihrmark Lundberg K, Ayyalusamy R, Baig MH, et al. METTL3/MYCN cooperation drives neural crest differentiation and provides therapeutic vulnerability in neuroblastoma. EMBO J. 2024 Dec;43(24):6310-35.
103. Sun Y, Shen W, Hu S, Lyu Q, Wang Q, Wei T, et al. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. J Exp Clin Cancer Res. 2023 Mar 17;42(1):65.
104. Cesaro B, Iaiza A, Piscopo F, Tarullo M, Cesari E, Rotili D, et al. Enhancing sensitivity of triple-negative breast cancer to DNA-damaging therapy through chemical inhibition of the m6A methyltransferase METTL3. Cancer Commun (Lond). 2024 Feb;44(2):282-6.
105. Fang M, Li Y, Wang P, Wang Y, Wang X, Wa X, et al. METTL3 Inhibition Restores PD-L1 Expression and CD8+ T-cell Cytotoxic Function in Immunotherapy-Treated Gastric Cancer. Cancer Immunol Res. 2025 Jul 2;13(7):1037-52.
106. Yu H, Liu J, Bu X, Ma Z, Yao Y, Li J, et al. Targeting METTL3 reprograms the tumor microenvironment to improve cancer immunotherapy. Cell Chem Biol. 2024 Apr 18;31(4):776-91.e7.
107. Wu L, Bai R, Zhang Y, Chen H, Wu J, Chen Z, et al. METTL3-VISTA axis-based combination immunotherapy for APC truncation colorectal cancer. J Immunother Cancer. 2024 Dec 9;12(12):e009865.
108. Tong Y, Chen Z, Wu J, Huang Q, He Y, Shang H, et al. METTL3 promotes an immunosuppressive microenvironment in bladder cancer via m6A-dependent CXCL5/CCL5 regulation. J Immunother Cancer. 2025 Apr 15;13(4):e011108.
109. Ou YJ, Liu BJ, Xuan YF, Bao XB, Huan XJ, Song SS, et al. The combination of BET and METTL3 inhibitors elicits synergistic antitumor effects in ovarian cancer cells via reducing SP1 and BCL-2 expression. Life Sci. 2025 May 1;368:123505.
110. Zhang ZW, Zhao XS, Guo H, Huang XJ. The role of m6A demethylase FTO in chemotherapy resistance mediating acute myeloid leukemia relapse. Cell Death Discov. 2023 Jul 5;9(1):225.
111. Lin Z, Wan AH, Sun L, Liang H, Niu Y, Deng Y, et al. N6-methyladenosine demethylase FTO enhances chemo-resistance in colorectal cancer through SIVA1-mediated apoptosis. Mol Ther. 2023 Feb 1;31(2):517-34.
112. Ren X, Tang X, Huang T, Hu Z, Wang Y, Zhou Y. FTO plays a crucial role in gastrointestinal cancer and may be a target for immunotherapy: an updated review. Front Oncol. 2023 Oct 12;13:1241357.
113. Zhang S, Zhou L, Lu J, Yang J, Tao L, Feng Y, et al. The FTO inhibitor Rhein is a promising option for the treatment of multidrug resistance AML. 2024. DOI: https://doi.org/10.21203/rs.3.rs-3813501/v1.
114. Dong Z, Huang Y, Xia W, Liao Y, Yang CG. A patenting perspective of fat mass and obesity associated protein (FTO) inhibitors: 2017-present. Expert Opin Ther Pat. 2025 Jun;35(6):533-42.
115. Chen A, Zhang VX, Zhang Q, Sze KM, Tian L, Huang H, et al. Targeting the oncogenic m6A demethylase FTO suppresses tumourigenesis and potentiates immune response in hepatocellular carcinoma. Gut. 2025 Jan 1;74(1):90-102.
116. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019 Jun 25;10(1):2782.